THE IDEA
Nerve cells in the brain establish connections or synapses to form complex electrical circuits that keep people thinking and moving. Despite the importance of these synapses in mediating the flow of charged particles between neurons, not much is understood about how these connections are created.


Research conducted by graduate student Sierra Palumbos and Professor of Cell and Developmental Biology David Miller sought to identify the underlying genetic mechanisms that specify the location of the brain’s electrical synapses.
Using the nematode C. elegans as a model, the researchers developed new methods to measure function of active electrical synapses called gap junctions, and they deployed live cell imaging techniques—developed in Miller’s lab—to visualize them. This is the first time that these techniques have been used in the intact nervous system of a living organism.
WHY IT MATTERS
“We have identified a signaling pathway that directs the formation of electrical synapses between specific neurons,” Palumbos said. A key component of this pathway, cyclic AMP or cAMP, drives the movement of gap junction components to specific destinations in each neuron for assembly into electrical synapses.
C. elegans is especially useful for these studies because its nervous system has only 302 neurons, compared with the human brain’s 100 billion. Also, human and C. elegans nervous systems are guided by similar genetic rules. Thus, research conducted in C. elegans offers an indirect but powerful strategy for illuminating the genetic instructions that build the human brain. Because cAMP is also found in human neurons, this work in C. elegans predicts that cAMP directs assembly of electrical synapses in the brain, a phenomenon that can be altered by stroke or neurodegenerative disease.
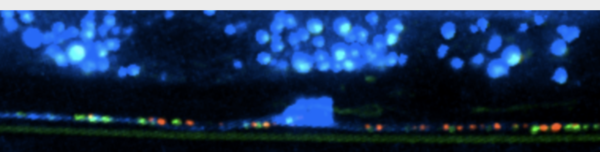
WHAT’S NEXT
Electrical synapses are composed of modular components that are routed to specific destinations in each neuron in the brain for assembly. The next step is to identify the molecular “motor” that transports these components and to establish how cAMP controls its trafficking activity, Miller said.
FUNDING
This work was supported by the American Heart Association, NIH grant R01NS113559 and NSF grant DGE:1445197. Experiments were performed in the Vanderbilt Flow Cytometry Shared Resource and in the Cell Imaging Shared Resource, which is supported by NIH grant DK020593.
GO DEEPER
The article, “cAMP controls a trafficking mechanism that maintains the neuron specificity and subcellular placement of electrical synapses during development” was published in the journal Developmental Cell on Nov. 5.
Other co-authors include former graduate students Steve Von Stetina and Rachel Skelton, summer undergraduate interns Isaiah Swann and Amanda Mitchell, and Harpeth Hall high school student Sydney Heifner. Heifner, a participant in an honors STEM research program, conducted her research in Miller’s lab. There she learned to use the confocal microscope to capture images of fluorescently labeled gap junctions in wild type and mutant backgrounds. Her work appears as a figure in this paper.