By applying unsupervised and automated machine learning techniques to the analysis of millions of cancer cells, Rebecca Ihrie and Jonathan Irish, both associate professors of cell and developmental biology, have identified new cancer cell types in brain tumors. Machine learning is a series of computer algorithms that can identify patterns within enormous quantities of data and get ‘smarter’ with more experience. This finding holds the promise of enabling researchers to better understand and target these cell types for research and therapeutics for glioblastoma – an aggressive brain tumor with high mortality – as well as the broader applicability of machine learning to cancer research.

With their collaborators, Ihrie and Irish developed Risk Assessment Population IDentification (RAPID), an open-source machine learning algorithm that revealed coordinated patterns of protein expression and modification associated with survival outcomes.
The article, “Unsupervised machine learning reveals risk stratifying glioblastoma tumor cells” was published online in the journal eLife on June 23. RAPID code and examples are available on the cytolab Github page.
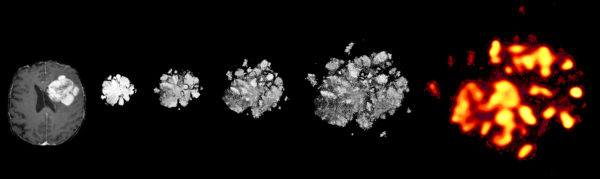
For the past decade, the research community has been working to leverage machine learning’s ability to absorb and analyze more data for cancer cell research than the human mind alone can process. “Without any human oversight, RAPID combed through 2 million tumor cells – with at least 4,710 glioblastoma cells from each patient – from 28 glioblastomas, flagging the most unusual cells and patterns for us to look into,” said Ihrie. “We’re able to find the needles in the haystack without searching the entire haystack. This technology lets us devote our attention to better understanding the most dangerous cancer cells and to get closer to ultimately curing brain cancer.”
Fed into RAPID were data on cellular proteins that govern the identity and function of neural stem cells and other brain cells. The data type used is called single-cell mass cytometry, a measurement technique typically applied to blood cancer. Once RAPID’s statistical analysis was complete and the “needles in the haystack” were found, only those cells were studied. “One of the most exciting results of our research is that unsupervised machine learning found the worst offender cells without needing the researchers to give it clinical or biological knowledge as context,” said Irish, also scientific director of Vanderbilt’s Cancer & Immunology Core. “The findings of this study currently represent the biggest biology advance from my lab at Vanderbilt.”
The researchers’ machine learning analysis enabled their team to study multiple characteristics of the proteins in brain tumor cells in relation to other characteristics, delivering new and unexpected patterns. “The collaboration between our two labs, the support that we received for this high-risk work from Vanderbilt and the Vanderbilt-Ingram Cancer Center (VICC) and the fruitful collaboration with neurosurgeons and pathologists who provided a unique opportunity to study human cells right out of the brain allowed us to achieve this milestone,” said Ihrie and Irish in a joint statement. The co-first authors of the paper are former Vanderbilt graduate students Nalin Leelatian, a current neuropathology resident at Yale (Irish lab), and Justine Sinnaeve (Ihrie lab). Through her research and work on this topic, Leelatian earned the American Brain Tumor Association (ABTA) Scholar-in-Training Award, American Association for Cancer Research (AACR) in April 2017.
The applicability of this research extends beyond cancer research to data analysis techniques for broader human disease research and laboratory modeling of diseases using multiple samples. The paper also demonstrates that these complex patterns, once found, can be used to develop simpler classifications that can be applied to hundreds of samples. Researchers studying glioblastoma brain tumors will be able to refer to these findings as they test to see if their own samples are comparable to the cell and protein expression patterns discovered by Ihrie, Irish, and collaborators.
This work was supported by the Michael David Greene Brain Cancer Fund, a discovery grant for brain tumor research established in 2004. The grant was recently renewed for another five years to support Ihrie and Irish’s continued research on glioblastoma. Additional support was provided by the National Institutes of Health, including the National Cancer Institute and National Institute of Neurological Disorders and Stroke, VICC and VICC Ambassadors, the Vanderbilt International Scholars program, a Vanderbilt University Discovery Grant, an Alpha Omega Alpha Postgraduate Award, a Society of Neurological Surgeons/RUNN Award, a Burroughs Wellcome Fund Physician-Scientist Institutional Award, the Vanderbilt Institute for Clinical and Translational Research, and the Southeastern Brain Tumor Foundation.