
Cocaine, nicotine, capsaicin.
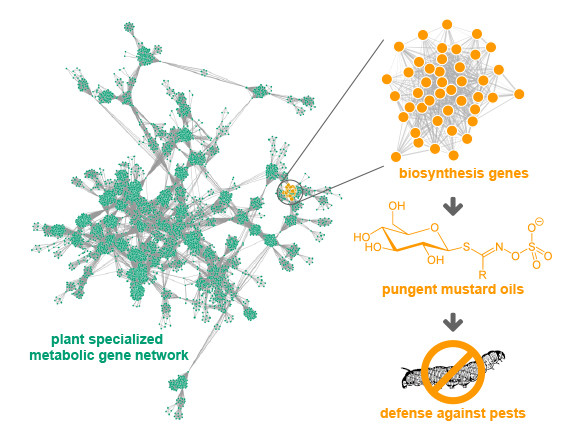
These are just three familiar examples of the hundreds of thousands of small molecules (also called specialized or secondary metabolites) that plants use as chemical ammunition to protect themselves from predation.
Unfortunately, identifying the networks of genes that plants use to make these biologically active compounds, which are the source of many of the drugs that people use and abuse daily has vexed scientists for years, hindering efforts to tap this vast pharmacopeia to produce new and improved therapeutics.

Now, Vanderbilt University geneticists think they have come up with an effective and powerful new way for identifying these elusive gene networks, which typically consist of a handful to dozens of different genes, that may overcome this road block.
“Plants synthesize massive numbers of bioproducts that are of benefit to society. This team has revolutionized the potential to uncover these natural bioproducts and understand how they are synthesized,” said Anne Sylvester, program director in the National Science Foundation’s Biological Sciences Directorate, which funded the research.
The revolutionary new approach is based on the well-established observation that plants produce these compounds in response to specific environmental conditions.
“We hypothesized that the genes within a network that work together to make a specific compound would all respond similarly to the same environmental conditions,” explained Jennifer Wisecaver, the postdoctoral fellow who conducted the study.
To test this hypothesis, Wisecaver – working with Cornelius Vanderbilt Professor of Biological Sciences Antonis Rokas and undergraduate researcher Alexander Borowsky – turned to Vanderbilt’s in-house supercomputer at the Advanced Computing Center for Research & Education in order to crunch data from more than 22,000 gene expression studies performed on eight different model plant species.
“These studies use advanced genomic technologies that can detect all the genes that plants turn on or off under specific conditions, such as high salinity, drought or the presence of a specific predator or pathogen,” said Wisecaver.
But identifying the networks of genes responsible for producing these small molecules from thousands of experiments measuring the activity of thousands of genes is no trivial matter. That’s where the Vanderbilt scientists stepped in; they devised a powerful algorithm capable of identifying the networks of genes that show the same behavior (for example, all turning on) across these expression studies.
The result of all this number crunching – described in the paper titled “A global co-expression network approach for connecting genes to specialized metabolic pathways in plants” published on April 13, 2017, in The Plant Cell journal – was the identification of dozens, possibly even hundreds of gene pathways that produce small metabolites, including several that previous experiments had identified.
Vered Tzin from Ben-Gurion University’s Jacoob Blaustein Institutes for Desert Research in Israel and Georg Jander from Cornell University’s Boyce Thompson Institute for Plant Research in Ithaca, NY, helped verify the predictions the analysis made in corn, and Daniel Kliebenstein from the Department of Plant Sciences at the University of California, Davis helped verify the predictions in the model plant system Arabidopsis.
The results of their analysis go against the prevailing theory that the genes that make up these pathways are clustered together on the plant genome. “This idea comes from the observation in fungi and bacteria that the genes that make up these specialized metabolite pathways are clustered together,” said Rokas. “In plants, however, these genes appear to be mostly scattered across the genome. Consequently, the strategies for discovering plant gene pathways will need to be different from those developed in the other organisms.”
The researchers argue that the results of their study show that this approach “is a novel, rich and largely untapped means for high-throughput discovery of the genetic basis and architecture of plant natural products.”
If that proves to be true, then it could help open the tap on new plant-based therapeutics for treating a broad range of conditions and diseases.
The research was funded by a National Science Foundation National Plant Genome Initiative (NPGI) Postdoctoral Research Fellowship to Wisecaver (IOS-1401682), as well as by National Science Foundation grants to Rokas (DEB-1442113) and Jander (IOS-1339237).