
First it was chess. Then it was Jeopardy.
Now computers are at it again, but this time they are trying to automate the scientific process itself.
An interdisciplinary team of scientists at Vanderbilt University, Cornell University and CFD Research Corporation, Inc., has taken a major step toward this goal by demonstrating that a computer can analyze raw experimental data from a biological system and derive the basic mathematical equations that describe the way the system operates. According to the researchers, it is one of the most complex scientific modeling problems that a computer has solved completely from scratch.

The paper that describes this accomplishment is published in the October issue of the journal Physical Biology and is currently available online. The work was a collaboration between John P. Wikswo, the Gordon A. Cain University Professor at Vanderbilt, Michael Schmidt and Hod Lipson at the Creative Machines Lab at Cornell University and Jerry Jenkins and Ravishankar Vallabhajosyula at CFDRC in Huntsville, Ala.
The “brains” of the system, which Wikswo has christened the Automated Biology Explorer (ABE), is a unique piece of software called Eureqa developed at Cornell and released in 2009. Schmidt and Lipson originally created Eureqa to design robots without going through the normal trial and error stage that is both slow and expensive. After it succeeded, they realized it could also be applied to solving science problems.
One of Eureqa’s initial achievements was identifying the basic laws of motion by analyzing the motion of a double pendulum. What took Sir Isaac Newton years to discover, Eureqa did in a few hours when running on a personal computer.
In 2006, Wikswo heard Lipson lecture about his research. “I had a ‘eureka moment’ of my own when I realized the system Hod had developed could be used to solve biological problems and even control them,” Wikswo said. So he started talking to Lipson immediately after the lecture and they began a collaboration to adapt Eureqa to analyze biological problems.
“[rquote]Biology is the area where the gap between theory and data is growing the most rapidly,” said Lipson. “So it is the area in greatest need of automation.[/rquote]”
The biological system that the researchers used to test ABE is glycolysis, the primary process that produces energy in a living cell. Specifically, they focused on the manner in which yeast cells control fluctuations in the chemical compounds produced by the process.

The researchers chose this specific system, called glycolytic oscillations, to perform a virtual test of the software because it is one of the most extensively studied biological control systems. Jenkins and Vallabhajosyula used one of the process’ detailed mathematical models to generate a data set corresponding to the measurements a scientist would make under various conditions. To increase the realism of the test, the researchers salted the data with a 10 percent random error. When they fed the data into Eureqa, it derived a series of equations that were nearly identical to the known equations.
“What’s really amazing is that it produced these equations a priori,” said Vallabhajosyula. “The only thing the software knew in advance was addition, subtraction, multiplication and division.”
Beyond Adam
The ability to generate mathematical equations from scratch is what sets ABE apart from Adam, the robot scientist developed by Ross King and his colleagues at the University of Wales at Aberystwyth. Adam runs yeast genetics experiments and made international headlines two years ago by making a novel scientific discovery without direct human input. King fed Adam with a model of yeast metabolism and a database of genes and proteins involved in metabolism in other species. He also linked the computer to a remote-controlled genetics laboratory. This allowed the computer to generate hypotheses, then design and conduct actual experiments to test them.
“It’s a classic paper,” Wikswo said.
In order to give ABE the ability to run experiments like Adam, Wikswo’s group is currently developing “laboratory-on-a-chip” technology that can be controlled by Eureqa. This will allow ABE to design and perform a wide variety of basic biology experiments. Their initial effort is focused on developing a microfluidics device that can test cell metabolism.
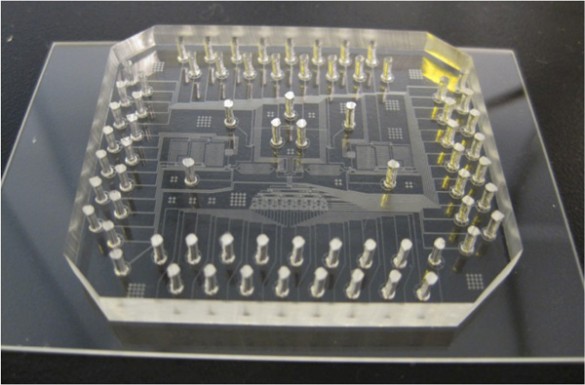
“Generally, the way that scientists design experiments is to vary one factor at a time while keeping the other factors constant, but, in many cases, the most effective way to test a biological system may be to tweak a large number of different factors at the same time and see what happens. ABE will let us do that,” Wikswo said.
The project was funded by grants from the National Science Foundation, National Institutes of Health, the Defense Threat Reduction Agency and the National Academies Keck Futures Initiative.
Why biology needs automation
“Biology is more complex than astronomy or physics or chemistry,” maintained Wikswo, a physicist who has spent his career studying biological systems. “In fact, it may be too complex for the human brain to comprehend.”
This complexity stems from the fact that biological processes range in size from the dimensions of an atom to those of a whale and in time from a billionth of a second to billions of seconds. Biological processes also have a tremendous dynamic range: for example, the human eye can detect a star at night that is one billionth as bright as objects viewed on a sunny day.
Then there is the matter of sheer numbers. A cell expresses between 10,000 to 15,000 proteins at any one time. Proteins perform all the basic tasks in the cell, including producing energy, maintaining cell structures, regulating these processes and serving as signals to other cells. At any one time there can be anywhere from three to 10 million copies of a given protein in the cell.
According to Wikswo, the crowning source of complication is that processes at all these different scales interact with one another: “These multi-scale interactions produce emergent phenomena, including life and consciousness.”
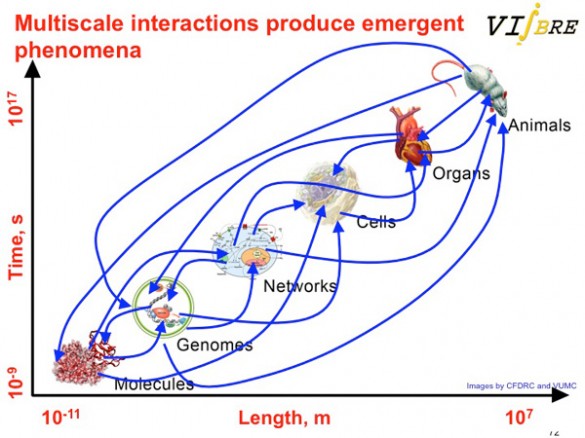
Looked at from a mathematical point of view, to create an accurate model of a single mammalian cell may require generating and then solving somewhere between 100,000 to one million equations.
Balanced against this complexity is the capability of the human brain. The biophysicist cites research that has found that the human brain can only process seven pieces of data at a time and quotes a 1938 assessment of brain research by Emerson Pugh: “If the human brain were so simple that we could understand it, we would be so simple that we couldn’t.”
That is where robot scientists like ABE and Adam come in, Wikswo argues. They have the potential for both generating and analyzing the tremendous amounts of data required to really understand how biological systems work and predict how they will react to different conditions.
Power of co-evolution
“We set out to work with robots, but our path took us, through many twists and turns, to automating science,” said Lipson, associate director of the Creative Machines Lab.
Lipson used a procedure called genetic programming for the breeding process. It involves starting with the basic components of a robot, randomly combining them in millions of different configurations and then testing how well they perform by a specific criterion, such as how fast they can move. The designs that work the best are then randomly combined and tested. These steps are repeated until it produces a design that is acceptable. However, this process also proved to be too slow.
So Lipson combined the breeding and the debugging processes in an approach he calls co-evolution. He started with a crude simulator, used it to design a robot, tested the design and studied how it failed. He used this information to improve the simulator so that it could predict the failure. Then he used the improved simulator to design another robot, tested the design, watched how it failed and improved the simulator once again. Repeating these steps of co-evolving simulators and robots produced increasingly competent designs, he found.
After proving that co-evolution works for robot design, Lipson realized that it could be generalized to solve other problems. Specifically, he adapted it for the mathematical process of curve fitting, more generally called symbolic regression. This involves deriving equations that can describe various data sets.
Lipson’s software package, which he and student Michael Schmidt named Eureqa, proved to be extremely successful. As the word got around, he began getting requests for copies of the program and decided to make it into a citizen science project, available for anyone to download on the Internet.
“Today, it has more than 20,000 users. People are using it to solve problems in a wide variety of areas including traffic, business and neighborhood problems,” Lipson said. He and his students tested it to see if they could predict the stock market, but it didn’t work. “It may have worked for others, who aren’t talking about it,” he added.
The software didn’t work on the first biology program it was given either. Gurol Suel, a researcher at the University of Texas Southwestern Medical Center, sent Lipson an extensive data set from his studies of single cell dynamics and asked him to run it through Eureqa. When Lipson and Schmidt did so and sent him back the results, Suel informed them they didn’t make any sense. As they thought about the problem, the researchers realized that they hadn’t given the software the tools it needed.
“We had given it the ability to add, subtract, multiply and divide and to calculate sines and cosines. But sines and cosines weren’t relevant, while other factors that we hadn’t included, such as time delays, were,” he explained. When they made this adjustment, Eureqa derived a set of elegant equations that were simpler than the ones Suel had derived, but Suel said that he didn’t know how to interpret them.
Understanding the meaning of the equations that Eureqa generates can be a problem, Lipson acknowledged: “We may have to create another program to do this.”
Wikswo isn’t as concerned. He maintains that this approach will give scientists the ability to control biological systems even if they can’t completely explain how they work, and this capability can provide the basis for the development of significantly improved drugs and other therapies.
Watch and learn
Video explaining how Eureqa derived the fundamental equations of motion from observations of a double pendulum.
One of Creative Machines Lab’s early efforts to breed robots that can move across different surfaces. After evolving virtual robots which can move, the researchers created physical versions of them using 3-D printing technology.