
Meet TrCel7a (pronounced tee-are-cell-seven-a).
TrCel7a is a cellulase: a special enzyme that breaks down cellulose, the most plentiful natural polymer on the planet.
The enzyme works like a microscopic wood chipper. It swallows strands of tightly bound cellulose and breaks them down into simple sugars. It works very slowly but, like a truck operating at a very low gear, it is extremely difficult to stop once it gets going. It is also self-propelling, powered in large part by energy from the cellulose bonds that it breaks.
Finding ways to make enzymes like TrCel7a operate faster and more efficiently could be the key to transforming ethanol made from cellulose into a major new renewable fuel source. In the U.S. each year an estimated 323 million tons of cellulosic wastes are thrown away – enough to provide as much as 30 percent of current fuel consumption.
“Until now, this system has been something of a black box at the molecular level. We knew what these enzymes did but we didn’t know how they worked,” said Matthew Lang, professor of chemical and biomolecular engineering at Vanderbilt University.
Working in the Lang Lab, doctoral student Sonia Brady has broken open this black box in the case of TrCel7A and looked inside. Borrowing a technique biophysicists use to study other molecular motors, she has measured the behavior of the enzyme and its constituent parts in unprecedented detail. The results of the study were published Dec. 10 in the online journal Nature Communications.
“Measuring the behavior of an individual enzyme and its component parts is a new strategy in the study of cellulose decomposition,” said Brady. “We hope that it will provide knowledge others can use to integrate biological strategies with industrial processes in new and exciting ways.”
For the last eight years, the U.S. government has been backing a major program to develop advanced biofuels. One of the main areas of research in cellulosic ethanol has been to find the most effective and low-cost microorganisms and enzymes to use in the process. Much of this effort has centered on finding ways to get cellulase-producing microorganisms to make larger amounts of these enzymes.
Brady and Lang, on the other hand, decided to study the way individual cellulase enzymes work. To do so, they selected a cellulase produced by the filamentous fungus Trichoderma reesei, one of the microorganisms used commercially to decompose cellulose. T. reesei produces a cocktail of three different enzymes that it uses for this purpose, 60 percent of which (by mass) is TrCel7A.
To get the first direct measurements of the mechanical properties of individual TrCel7A molecules, the researchers used an instrument called optical tweezers, which grasp and manipulate extremely small objects with a laser beam. They don’t work on anything as small as an enzyme, however. So the researchers had to attach tiny polystyrene spheres to individual molecules. (Imagine a person holding the tether of one of the balloons in the Macy’s Thanksgiving Day Parade.)
Once such a sphere is attached and the enzyme is placed on a cellulose fiber, it drags the sphere behind it as it works. This allowed the researchers to track its otherwise invisible movements. Equally as important, the researchers could grab the sphere with the optical tweezers and exert force on it…force that is transferred to the enzyme. By assisting and resisting the enzyme’s motion, the researchers determined how much force it takes to slow TrCel7A down and how much it speeds up when it is pulled along.
The researchers discovered that the enzyme is very slow. It creeps along a cellulose strand at an average of only 0.25 nanometers per second. That’s about the width of 10 hydrogen atoms per second. And they found that its movement consisted of an alternation of one-nanometer steps and dwell times of varying lengths.
“It doesn’t look as if it’s very easy to increase the step length because it appears to be directly related to the length of the glucose units in the cellulose that it is breaking apart,” said Lang. “However, when we applied an assisting force we could double to enzyme’s velocity by reducing the dwell times.”
In addition, the researchers broke down the enzyme into its component parts and clarified the role that each of the parts play.
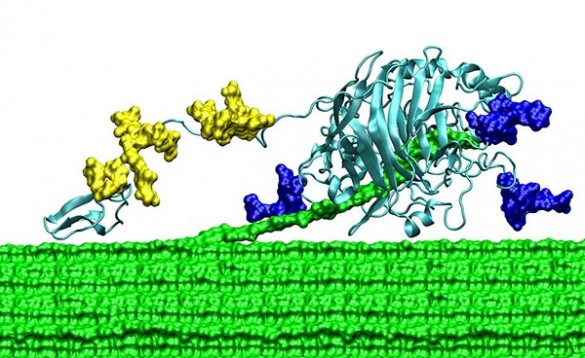
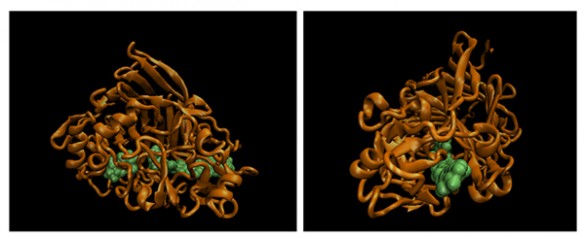
TrCel7a has three basic components: a main “catalytic domain (CD),” a much smaller “carbohydrate-binding module (CBM)” and a flexible “linker domain (LD)” that connects the two.
“We call the CBM and LD the CD’s ball and chain,” said Lang.
Using the enzyme’s genetic blueprint, the researchers made versions of the catalytic domain without the LD and CBM attached. They discovered that once this naked CD found a strand of cellulose to chew on, it behaved in an identical fashion to the whole enzyme.
“This raised the question of why the CD keeps its ball and chain,” Lang said.
The answer seems to be that by attaching to cellulose fiber, the CBM holds the CD in the immediate vicinity of the fiber so it can find loose ends that it can ingest more readily. Their studies show that the rate at which CD without its ball and chain initiates processing of cellulose is only 1/50th that of the whole enzyme.
One of the limitations of TrCel7A is that it needs to find a broken end of a cellulose strand to begin working. That is the reason T. reesei produces another type of enzyme called an endocellulase. This takes short chunks of cellulose fibers, essentially creating loose ends that TrCel7A can begin digesting. As a result, the decomposition rate of the two enzymes working together is substantially higher than what they can achieve working independently.
According to Lang, the next step in their research is to study how ensembles of these enzymes work together at the molecular level.
Coauthors on the study were Vanderbilt doctoral student Yinnian Feng, Sreelatha Sarangapani at the National University of Singapore and Shishir Chundawat at Rutgers, The State University of New Jersey.
The research was funded by National Science Foundation grants 1330792 and 1236120, U.S. Department of Energy grant DE-FC02-07ER64494 and U.S. Department of Education Graduate Assistance in Areas of National Need grant P200A090323 and the Singapore-MIT Alliance for Research and Technology-BioSyM.