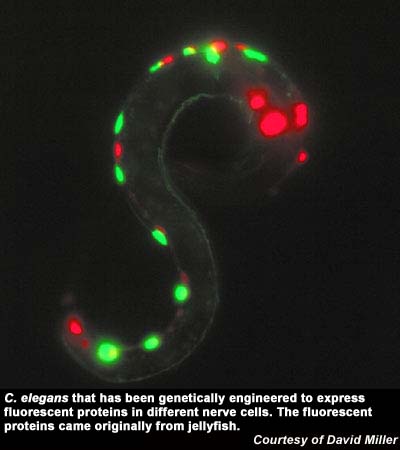
 The roundworm C. elegans provides a simple yet representative model of development that depends on differential expression of a compact, well-described genome. Although the developmental origin of each C. elegans cell is fully described, a comparable gene expression map is not currently available.
The roundworm C. elegans provides a simple yet representative model of development that depends on differential expression of a compact, well-described genome. Although the developmental origin of each C. elegans cell is fully described, a comparable gene expression map is not currently available.
David Miller and colleagues have now produced a gene expression atlas for more than 30 different cell types and developmental stages. These results revealed that most worm genes are highly regulated across developmental stages and cell types. The researchers also detected a large number of novel genes of unknown function that comprise about 10 percent of the C. elegans genome.
The resulting spatial and temporal gene expression maps, reported in the February issue of Genome Research, provide a basis for establishing roles for individual genes in the development of specific cell types.
View the online resources the researchers established to facilitate the use of these data for future studies.