A zebrafish model of a rare genetic disease has revealed an unexpected regulatory pathway for cholesterol absorption and processing.
The findings, reported in the Journal of Molecular Medicine, were “quite surprising and have the potential to teach us about the basic physiology of cholesterol management,” said Ela Knapik, M.D., associate professor of Medicine and Cell and Developmental Biology.

Knapik and her colleagues were interested in the protein Sar1b — part of the transport machinery inside cells. They knew that mutations in the human Sar1b gene are associated with Anderson disease, a lipid malabsorption syndrome.
The investigators developed a zebrafish model of Sar1b deficiency to study roles the protein might play during development. The most striking finding, Knapik said, was that the zebrafish missing Sar1b did not absorb or process cholesterol. Patients with Anderson disease have low levels of cholesterol in the blood, but this had been attributed to poor absorption of lipids and other nutrients — not to defects in cholesterol absorption.
“If we can learn how Sar1b helps regulate intracellular cholesterol, it may give us clues about how to better manage cholesterol levels in various disease states,” Knapik said.
The zebrafish missing Sar1b retained lipids (fats) inside intestinal cells, resembling the characteristics of patients with Anderson disease. The fish also had defects in the growth and development of the pancreas and liver, similar to patients with Anderson disease.
“Exocrine pancreas deficiency is a very common symptom in patients, and it has been presumed to be secondary to poor lipid absorption,” Knapik said. “In our model, we see these deficits in pancreas and liver before the fish eat, suggesting that there are developmental deficits that add to the symptoms in patients.”
Knapik suggested that CT or ultrasound imaging to check the pancreas and liver of children with inherited mutations in Sar1b could impact diagnosis and clinical management for these patients.
“The more we know, the more clever we can be in diagnosing and treating this disease,” she said.
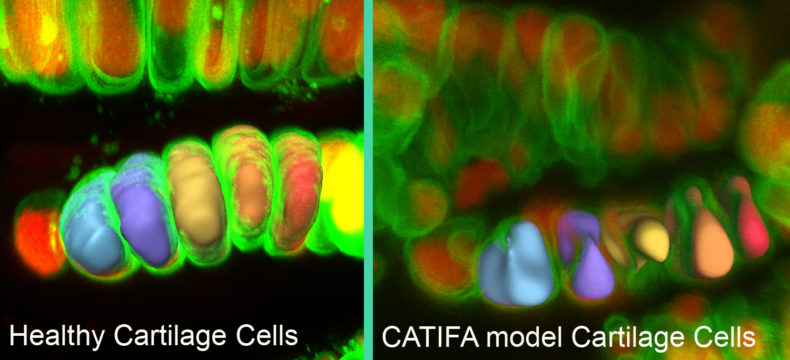
The zebrafish also had deficits in craniofacial cartilage formation and development of some neuronal populations.
Knapik’s team demonstrated that the human Sar1b gene could “rescue” the deficits in the zebrafish model. They are now developing a model where they can substitute the human Sar1b gene containing reported patient mutations to discover whether certain mutations affect specific developmental pathways.
“We want to understand the pathophysiology of the disease,” Knapik said. “And when we have these models, we can use them as screening tools for drugs to treat specific symptoms.”
Daniel Levic, a graduate student in Knapik’s laboratory, led the studies. Other participants in the research included J.R. Minkel, Wen-Der Wang, Ph.D., Witold Rybski and David Melville, Ph.D. The research was supported in part by the Zebrafish Initiative of the Vanderbilt University Academic Venture Capital Fund and by a grant from the National Institutes of Health (DE018477).