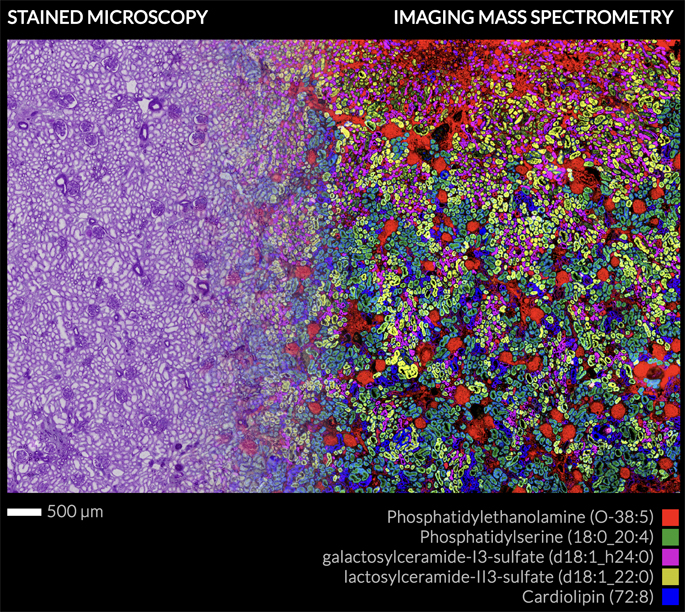
MALDI imaging mass spectrometry (IMS) is a powerful tool for studying the spatial distribution of molecules directly within tissues. It has been applied to many different pharmaceuticals, metabolites and small, soluble proteins. Large membrane-spanning proteins, however, have not been well studied by MALDI IMS because of analytical challenges related to their size and solubility.
Richard Caprioli, Ph.D., Stanford Moore Professor of Biochemistry, and colleagues have now developed a novel sample preparation procedure for MALDI IMS of transmembrane proteins directly from tissue. They used the method to assess the spatial distribution of myelin proteolipid protein and DM-20, two highly abundant transmembrane proteins within central nervous system myelin, throughout various regions of the rat brain. The study also includes new processes for assessing fatty acid modifications and for on-tissue protein identification using a hydrogel containing proteases (protein “scissors”).
The approaches, reported in the Aug. 6 issue of Analytical Chemistry, will enable further MALDI IMS studies of transmembrane protein distribution.
This research was supported by a grant from the National Institutes of Health (GM103391).